NSF funded research (DMS-0920887)
Double stranded (Ds) DNA viruses such as bacteriophages, adenoviruses and herpesviruses keep their genome in a spherical protein container called a capsid. The radius of the capsid is at least two orders of magnitude smaller than the length of the viral genome. This implies that once the DNA is packed inside the capsid it is subjected to strong bending and repulsive interactions (due to its rigidity and negative charge). Little is currently understood about the biophysical properties of the DNA molecule under these extreme conditions. In our group we use bacteriophage P4 to investigate such properties.
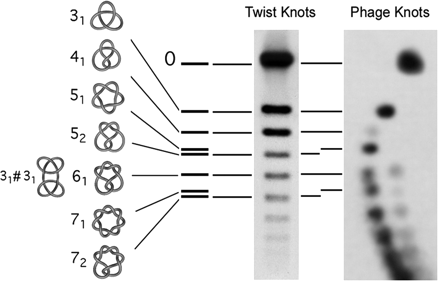
Bacteriophages, viruses that propagate in bacteria, are commonly used to study DNA packing and folding in other dsDNA viruses because they have similar morphology and share similar assembly pathways. A number of models have been proposed to explain the folding of the DNA molecule inside the bacteriophage capsid. However these models provide only a very general description of the trajectory of the DNA. This lack of understanding is reflected in the inability of current DNA folding models to predict the finding that DNA molecules abruptly extracted from bacteriophage P4 are knotted (i.e. are circles which cannot be laid flat on a plane without self-intersecting).
In our work, we have experimentally shown that P4 knots preserve information about the folding of the DNA inside the capsid and about the physical properties of the DNA itself. We are currently developing a model of DNA folding which improves on the models now available by incorporating biophysical properties that account for and allow for the reproduction of the knotted structures observed in P4. The long-term goal of this work is to provide a novel quantitative description of the process of DNA packing and folding inside dsDNA viruses that is consistent with the topological information observed in P4 and with other published data.