Work funded by NSF CAREER Award DMS1519375
Multiple cellular processes such as replication, recombination, and packing change the topology of DNA. Controlling these changes is key to ensuring stability inside the cell. The cell uses enzymes to simplify DNA topology. Techniques from knot theory and low-dimensional topology, aided by computational tools, now make it possible for us to study the specific action of such enzymes in these processes. Of special interest is the study of unlinking of DNA replication links by site-specific recombination. We model site-specific recombination as a local reconnection event and study the resulting reconnection pathways. This work is also relevant in fluid dynamics.
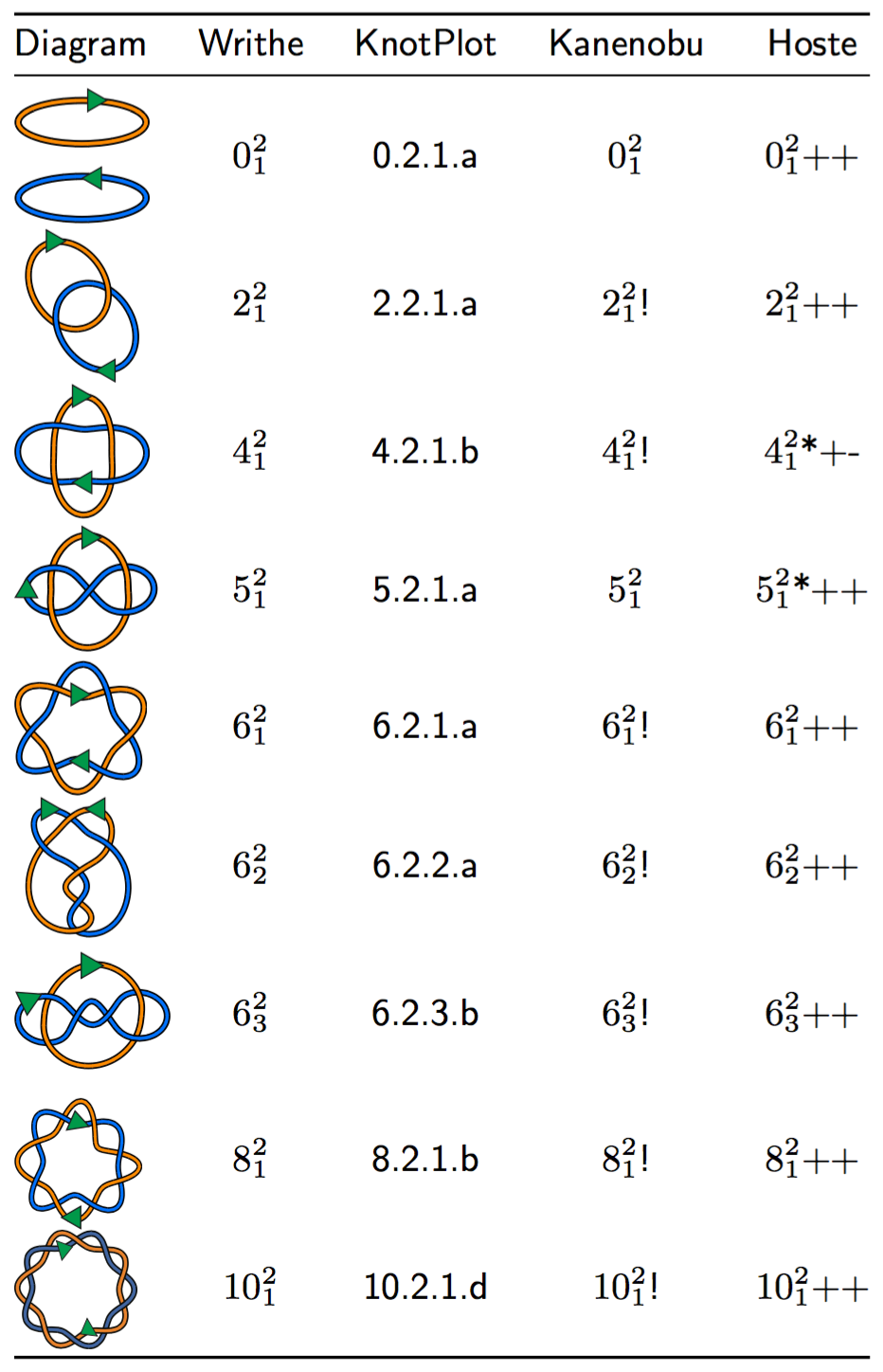
Link Table with nomenclature comparisons to other known tables.
See Knot Tables s5 s6 for a table of all of the knots with 10 or fewer crossings with specification of the chirality, according to the writhe-guided naming convention proposed in Brasher et al., 2013. . Knots in yellow have the same chirality as in Rolfsen’s Knot Table in his book Knots and Links, and those in red have chirality opposite to those of Rolfsen’s.